Abstract
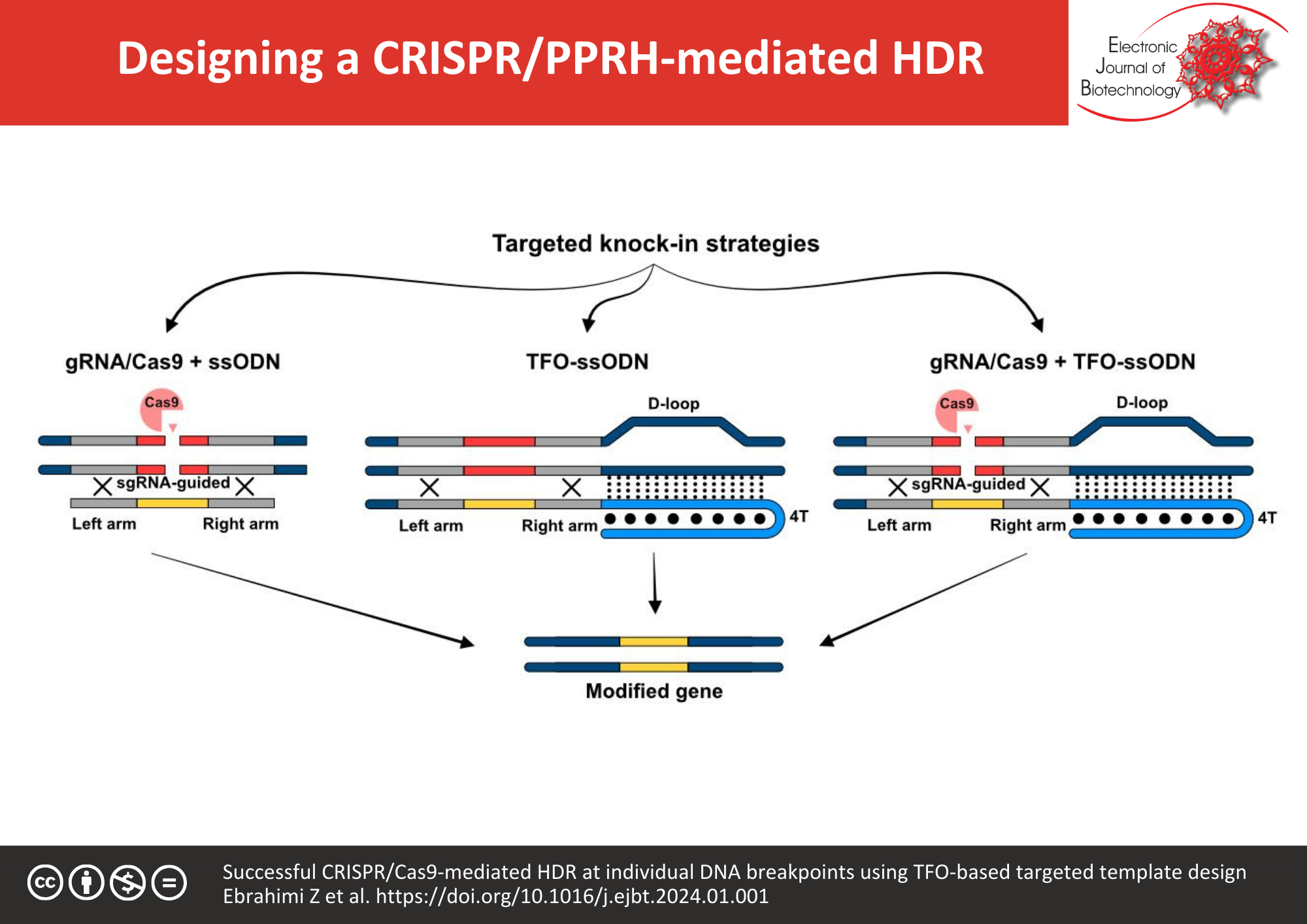
Background: Targeted insertion of the repair template into the genome is a common strategy for high-precision base replacements; however, the main challenge likely remains regarding the limited efficiency of homologous-directed repair (HDR). A precise genome cut achieved by CRISPR-Cas9 system combining with a single-stranded oligodeoxynucleotide (ssODN), as the donor template, improves significantly the rate of HDR. It is well-established that the spatial availability of the donor template to the repair system effectively enhances knock-in events in CRISPR-Cas9. PolyPurine Reverse Hoogsteen hairpins (PPRHs), as an alternative repairing strategy, benefits from a Triplex-forming oligonucleotide (TFO) for the repair template providing the ease of access. The main objective of the study was to evaluate the HDR frequency as a result of improvement of the spatial accessibility of the donor template adjacent to the cutting site. Hence, a flanking purine-rich hairpin complementary to the genomic DNA adjacent to the repairing site was fused to the ssODN with the incorporated bases for the alteration of EGFP to EBFP.
Results: Results from the comparison between the donor templates, ssODN and TFO-tailed ssODN, demonstrated an increased rate of knock-in from 18.2% ± 1.09 to 38.3% ± 4.54, respectively. From another perspective, findings indicated that the targeted Cas9-mediated DNA cleavage improves the efficiency of the repair-PPRH approach four-fold, as well.
Conclusions: The present study provides a viewpoint that highlights the significance of the designing of the donor template in terms of the structural features and positional access for the HDR-based repairing CRISPR-Cas9 systems.
References
Najafi S, Tan SC, Aghamiri S, et al. Therapeutic potentials of CRISPR-Cas genome editing technology in human viral infections. Biomedicine & Pharmacotherapy 2022;148:112743. https://doi.org/10.1016/j.biopha.2022.112743 PMid: 35228065
Schubert MS, Thommandru B, Woodley J, et al. Optimized design parameters for CRISPR Cas9 and Cas12a homology-directed repair. Scientific Reports 2021;11(1):19482. https://doi.org/10.1038/s41598-021-98965-y PMid: 34593942
Miyaoka Y, Berman JR, Cooper SB, et al. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Scientific Reports 2016;6(1):23549. https://doi.org/10.1038/srep23549 PMid: 27030102
Kurihara T, Kouyama-Suzuki E, Satoga M, et al. DNA repair protein RAD51 enhances the CRISPR/Cas9-mediated knock-in efficiency in brain neurons. Biochemical and Biophysical Research Communications 2020;524(3):621-8. https://doi.org/10.1016/j.bbrc.2020.01.132 PMid: 32029273
Li X-L, Li G-H, Fu J, et al. Highly efficient genome editing via CRISPR–Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Research. 2018;46(19):10195-215. https://doi.org/10.1093/nar/gky804 PMid: 30239926
Wang Y, Liu KI, Sutrisnoh N-AB, et al. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biology 2018;19:62. https://doi.org/10.1186/s13059-018-1445-x PMid: 29843790
Aird EJ, Lovendahl KN, St Martin A, et al. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Communications Biology 2018;1:54. https://doi.org/10.1038/s42003-018-0054-2 PMid: 30271937
Savic N, Ringnalda FC, Lindsay H, et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife 2018;7:e33761. https://doi.org/10.7554/eLife.33761 PMid: 29809142
Butt H, Eid A, Ali Z, et al. Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Frontiers in Plant Science 2017;8:1441. https://doi.org/10.3389/fpls.2017.01441 PMid: 28883826
Rodríguez L, Villalobos X, Dakhel S, et al. Polypurine reverse Hoogsteen hairpins as a gene therapy tool against survivin in human prostate cancer PC3 cells in vitro and in vivo. Biochemical Pharmacology 2013;86(11):1541-54. https://doi.org/10.1016/j.bcp.2013.09.013 PMid: 24070653
Noé V, Ciudad CJ. Polypurine reverse-hoogsteen hairpins as a tool for exon skipping at the genomic level in mammalian cells. International Journal of Molecular Sciences 2021;22(7):3784. https://doi.org/10.3390/ijms22073784 PMid: 33917446
de Almagro MC, Coma S, Noé V. Polypurine hairpins directed against the template strand of DNA knock down the expression of mammalian genes. Journal of Biological Chemistry. 2009;284(17):11579-89. https://doi.org/10.1074/jbc.M900981200 PMid: 19261618
Félix AJ, Ciudad CJ, Noé V. Correction of the aprt gene using repair-polypurine reverse Hoogsteen hairpins in mammalian cells. Molecular Therapy-Nucleic Acids. 2020;19:683-95. https://doi.org/10.1016/j.omtn.2019.12.015 PMid: 31945727
Albadri S, Del Bene F, Revenu C. Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 2017;121-122:77-85. https://doi.org/10.1016/j.ymeth.2017.03.005 PMid: 28300641
Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proceedings of the National Academy of Sciences 1994;91(26):12501-4. https://doi.org/10.1073/pnas.91.26.12501 PMid: 7809066
Félix AJ, Solé A, Noé V, et al. Gene correction of point mutations using polypurine reverse Hoogsteen hairpins technology. Frontiers in Genome Editing. 2020;2:583577. https://doi.org/10.3389/fgeed.2020.583577 PMid: 34713221
Boel A, De Saffel H, Steyaert W, et al. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Disease Models & Mechanisms 2018;11(10):dmm035352. https://doi.org/10.1242/dmm.035352 PMid: 30355591
Chen K, Gao C. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Reports 2014;33:575-83. https://doi.org/10.1007/s00299-013-1539-6 PMid: 24277082
García JF, Reguera D, Valls A, et al. Detection of pyrimidine-rich DNA sequences based on the formation of parallel and antiparallel triplex DNA and fluorescent silver nanoclusters. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2023;297:122752. https://doi.org/10.1016/j.saa.2023.122752 PMid: 37084680
Liang X, Potter J, Kumar S, et al. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. Journal of Biotechnology. 2017;241:136-46. https://doi.org/10.1016/j.jbiotec.2016.11.011 PMid: 27845164
Mitas M, Yu A, Dill J, et al. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat:(CTG) 15. Nucleic Acids Research 1995;23(6):1050-9. https://doi.org/10.1093/nar/23.6.1050 PMid: 7731793
Solé A, Ciudad CJ, Chasin LA, et al. Correction of point mutations at the endogenous locus of the dihydrofolate reductase gene using repair-PolyPurine Reverse Hoogsteen hairpins in mammalian cells. Biochemical Pharmacology. 2016;110-111:16-24. https://doi.org/10.1016/j.bcp.2016.04.002 PMid: 27063945
Solé Ferré A, Félix AJ, Noé Mata V. Polypurine Reverse Hoogsteen Hairpins as a tool for gene repair and editing. Recent Advances in Pharmaceutical Sciences VII, 2017, Research Signpost Editors: Diego Muñoz-Torrero, Montserrat Riu & Carles Feliu ISBN: 978-81-308-0573-3 Chapter 4, p 51-68. 2017.
Noé V, Aubets E, Felix AJ, et al. Nucleic acids therapeutics using PolyPurine Reverse Hoogsteen hairpins. Biochemical Pharmacology 2021;189:114371. https://doi.org/10.1016/j.bcp.2020.114371 PMid: 33338475
Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019;576(7785):149-57. https://doi.org/10.1038/s41586-019-1711-4 PMid: 31634902

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright (c) 2024 Electronic Journal of Biotechnology